Traits of good data hygiene and organization in Life Science R&D
This is the third post in a series focused on good data practices in Life Sciences. Here, we recap some specific traits of proper data hygiene and organization, with examples of what that looks like in practice. You can also check out our previous posts on why you should care about data and why people and culture are critical to the equation. If you find the stuff we're writing about or work we're doing at Kaleidoscope interesting, please reach out!
Summary points
- Maximize your impact. A lot of the practical steps to good data stewardship are, in essence, ways to maximize the value of any given piece of work over time. By being thoughtful about things like readability, discoverability, and documentation, you're ensuring that other team members (and future you) can benefit from the hard work you've done.
- Build alignment and shared understanding. Often, you can avoid (significant) wasted resources or expensive overhauls by setting a small amount of time aside to get team members on the same page. Even aligning on simple things like naming conventions or what tool to use for collaborative projects can make a big difference in how smoothly you run or how quickly you iterate.
- Start simple, get going, evolve over time. You don't have to start by implementing a system that will last you a decade; align on the immediate, low-hanging fruit and build from there. These initial steps can be low lift and low cost, and the earlier you start, the easier it will be, and the sooner you'll benefit from the quickly compounding gains.
When it comes to excellent data stewardship, you need to know what you’re striving for, to effectively rally your team and create a culture that cares about data. More specifically, when you’re first setting out to embed good data practices, it’s important to understand what the traits of well-maintained and organized data are, how those might differ across teams in your company, and which ones you want to optimize for. This way, you can chart a path between where you are today and where you want to get to, in order to start leveraging your growing body of data in the way that makes most sense for your organization.
Although this is by no means an exhaustive list, here are the more common traits of good data hygiene and organization that have been emphasized by the biotech companies we’ve spoken with.

Readability
Readable work is work that can be understood by yourself and by the people around you. Regardless of the form your work takes - code, documents, summaries of projects, or notes about lab procedures - it should be written in a way that makes it easy for someone else to read, understand, and distill key information from. For example, if your work consists of coding, making it readable might mean abstracting dense blocks of bloated code in notebooks by putting that code into functions and function libraries. For summaries of projects, this might mean taking a few moments to jot down a note that’s external to you – a note that someone else can read to quickly and effectively understand the context of the work you’re doing.
Making your work readable doesn’t mean that everything you write has to be polished to perfection – but it does mean making an effort to make your work as intuitive as possible so that it’s easier to understand, both for your future self and for others.
Version control and auditability
For those familiar with code, the well-known advice is to use source control to track all changes via Git or Github – but version control stretches beyond code, too. It’s about having a system where you can robustly track your work and its multiple iterations, regardless of file type – programs, web pages, even grant proposals. This way, you have a single source of truth that everyone can work from.
For some companies, the first step towards having a version-control system could be as simple as aligning on what core tools to use. At Kaleidoscope, we’ve spoken with multiple biotechs where each team uses a different product or tool depending on what they’re comfortable with. For example, when it comes to managing projects and progress towards milestones, one team will use Jira, another will use Trello, or Asana, or Clickup, and so on. In the end, this proliferation of tools that are unable to speak to each other results in work splintering across different functions and, all of a sudden, there’s no visibility. People can’t search across tools to see who did what, and no one can track what work is going on in teams other than their own.
These challenges become even more pronounced in contexts where a biotech is trying to bring together wet and dry labs more effectively or create a closed-loop system. Broadly speaking, these companies might have three groups working on different pieces of the puzzle: lab scientists generating experimental data, bioinformaticians analyzing experimental data, and engineers working on translating those learnings to deployed solutions. If every team is using different tools even when it’s for the same purpose (e.g. tracking a project), you’re left with unidirectional sharing of data and disparate environments, where each team throws their data ‘over the wall’ to the next team. In turn, this limits how quickly you can iterate and learn across functions – for instance, how easily lab scientists and bioinformaticians can discuss an analysis, or how quickly engineers can get feedback on something they’ve built related to that analysis. With a thoughtful system for holistically tracking iterations of work, people have top-level visibility into what work is being done and by who – making collaboration efficient and mistakes easier to clean up. Quick note: having a single system where it is theoretically possible to search across all work, does not mean that all work has to be visible to everyone. Good software tools should enable your team to have control over permissions on who has access to what.
Robustness: testing and code review
These traits are more specific to code, although the principles behind them can be applied to other types of work.
Testing means writing code that, in turn, makes sure your code works and does what you think it’s doing, and that it doesn’t do something like flip two columns of data and completely invert your findings. The expectation here shouldn’t be to mirror how sophisticated software engineers test code. For example, you don’t have to incorporate anything extreme like test-driven development, where you write the tests before you write the code. Rather, start with unit tests for only the mission-critical functions. In the process, you’ll catch bugs that will save you the headache of posthumously dissecting hundreds, if not thousands, of lines of code trying to figure out what went wrong.
Code reviews mean having multiple sets of eyes on your code or work to help catch mistakes. Rather than scheduling occasional formal reviews, you can institute more informal reviews and pair programming. This not only puts more eyes on more code, but facilitates knowledge exchange between the people on your team. Peer review is also relevant outside the realm of coding. Encourage people to regularly explain their work to other people on the team. Often, a fresh perspective is more apt at catching mistakes than the person who’s spent many hours or days on the same task.
Documentation (and standardization)
A large part of making sure your work is accessible is having well-thought out and user-friendly documentation throughout your organization.
For teams tasked with coding and analysis, this means having thorough documentation for functions, packages, and overall environment. Beyond code, there should be clear alignment on basic conventions like naming systems, tags, and formats (for example, deciding to format your dates like MM/DD/YYYY). Often, companies we speak with spend a lot of time integrating data that comes in different formats from different sources – lab machines, CROs, different partners – because they didn’t have this standardization figured out early enough. Don’t assume that something should be obvious and expect people to intuitively know how to label or call something. The goal post should be whether, in six months' time when someone new joins the team, they would have all the tools at their disposal to figure out what work was done and why.
This is also where metadata, or the data about the data, comes into play. Whether you’re doing wet lab or dry lab work, capturing the context of the work you’re doing – be it structural (where is the work stored), descriptive (who authored the document, when and why was that data collected) or administrative (what is the file type) – allows people to connect the dots between the what and why behind the science of any given experiment, project, or workflow.
At scale, this becomes powerful. Imagine legacy datasets sitting on servers, collecting dust – how can they be used without knowing when the data was collected or what the data means? In order for that data to tell meaningful stories and continue being valuable, it must be properly documented. The ability to retrieve, reuse, and track the data you generate so that it becomes useful beyond just today, is greatly enhanced when you create an environment where capturing the metadata about the science is just as important about the science itself. And while this is more of a future-proofing exercise rather than one for immediate utility, proper documentation of things like metadata shouldn’t feel like a distraction; it’s a relatively quick and painless task, once the team has aligned on the right system to use.
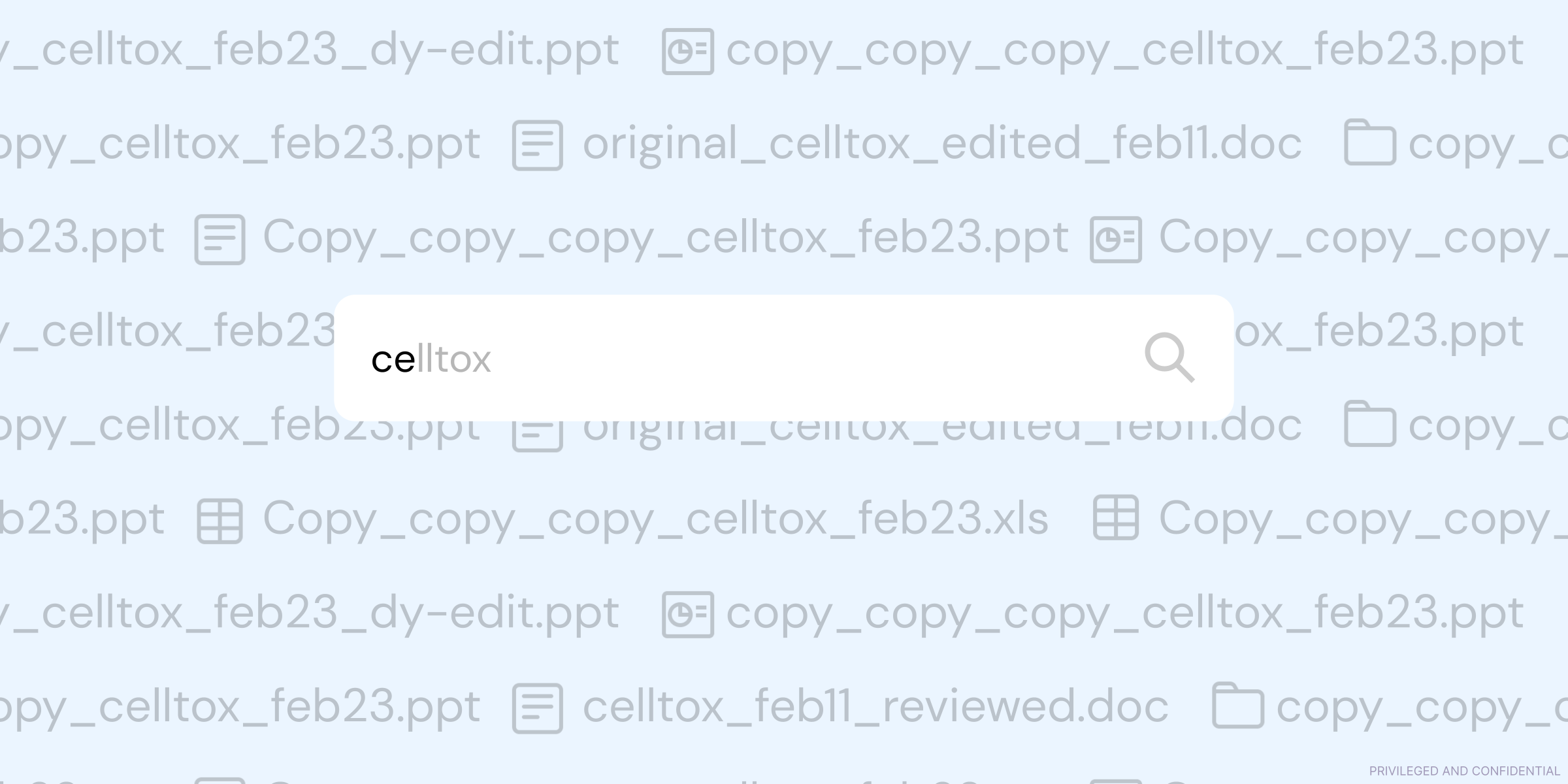
Discoverability
All the data you generate should be easy to find. Use a centralized repository that lets everyone in the organization access the data they need. This could be a server, a cloud-based system, or a data management platform – the point is that not knowing where something is should never be a blocker for anyone on your team. Again, standardization is important here. Plan a logical, consistent, and scalable directory structure that makes it clear how data should be saved and stored within folders and sub-folders, for example.
Having this mindset early on saves you from convoluted overhauls in your infrastructure. When you’re running or joining a small company, it can be tempting to use the easiest solution to get up and running – for example, everyone saving files to their own Google Drive folder. You might have the intention of centralizing everything “later” but, in reality, whatever solution you choose starts to grow deep roots. You’ll rely on elements of that infrastructure, customers will rely on that file system, you’ll keep reference data there, and so on. By the time you eventually do need to make the switch to a more robust system, there’ll be silos of information you’ll have to actively and painstakingly break down. If you promote discoverability from day one, the adaptations or upgrades you make along the way won’t feel nearly as jarring or disruptive to your team.
Securing your data against loss
This means storing your data in a secure location that is regularly backed up. The amount of biotech companies that still don’t do this is surprising. In 2022, the FDA sent out 56 violations to companies that failed to back-up their data.Without a backup and disaster recovery plan, your data is hostage to potential human error, hardware failure, virus attacks, power failure and more – and you’re setting yourself up for a future compliance nightmare when you have to start liaising with the FDA.
Having a data management plan
Thinking about incorporating all of these things might sound tedious but they can be tackled efficiently if you take the time to proactively establish and adopt a data management plan that everyone is familiar with. This plan should help you answer questions like:
- How will you collect data?
- How will you securely store and backup the data?
- Who will have access to what?
- Who is responsible for analyzing, archiving, and maintaining data?
- How will you train new members on these data management practices?
- What’s the lifecycle of your data (from how and when new data is created to when (if ever) it can be deleted)?
The upfront time and effort spent doing this might feel like a drag, but putting together a standardized plan from day one pays dividends in the long run – collaboration is easier, knowledge transfer is smoother, it becomes easier to spot errors in data, and regulatory audits are less stressful. Upfront effort is also always the lowest the earlier you are in your company’s life; you can align on what practices you want to follow and you don’t have as much of a backlog of mess to deconvolute.
You might come up with slightly different traits to the ones above – the important thing is that you set some time aside to write out what your benchmarks for good data hygiene and organization are, set checkpoints for achieving those benchmarks, and work backward from there. By seeing the gap between where you are now and where you want to be, you can come up with ways to close it and, in six months, you can look back and map the actions you took with the improvements they caused.
It’s worth noting that closing these gaps doesn’t mean building complex in-house systems; the gaps you have in your data management processes will also be ones that other biotechs feel or have felt, and it’s worth spending time looking into existing approaches, solutions, or software tools that can largely improve the way that you do things. Hopefully these improvements will help you create a new environment – one that no longer optimizes for the quick-and-dirty route, but instead produces work that is more reliable, more interpretable, and easier to extend and build upon.
If you want to chat more about anything we wrote, or you’re interested in finding a way to work together, let us know!
