Beyond Algorithms: Software Infrastructure for AI/ML Biotechs
Many of today's biotechs look nothing like yesterday’s. Companies can now run thousands of experiments in parallel, apply machine learning to predict molecular behavior, and process more data in a day than their predecessors did in a year. This shift brings a new set of challenges – not in the science itself, but in the software needed to support it.
In this post, we explore four critical challenges that AI/ML biotechs face - data generation & management, dataset preparation, exception handling, and data collection coordination - and the software solutions needed to address them. These aren't theoretical problems; they're the practical hurdles we've seen companies struggle with repeatedly.
Data Generation & Management
AI-driven biotechs produce massive datasets. To be useful, this data must be accurately annotated, stored with complete lineage, and queryable across experimental conditions. This is much harder than it sounds. Experimental data in biotech isn't just simple numbers. It's high-dimensional information with interconnected variables like sample metadata, experimental conditions, and references to previous results.
This complexity creates three challenges:
First, instrument metadata limitations. Many lab instruments (like NGS sequencers and imaging systems) strip or truncate important metadata, forcing scientists to create manual workarounds that are error-prone and time-consuming. We've seen teams spend hours reconstructing experiment contexts that should have been automatically captured.
Second, fragmented databases. Experimental results often end up stored in multiple systems that don't talk to each other, making lineage tracking nearly impossible. A scientist might need to check an electronic lab notebook for protocol details, an instrument database for raw results, and a separate analysis platform for processed data – all to understand a single experiment.
Third, the combinatorial explosion of experimental conditions. A relatively simple study might test five concentrations of three compounds across four cell lines with two timepoints. That's 120 distinct conditions, each generating multiple data points. Standard databases simply weren't designed to handle this dimensional complexity.
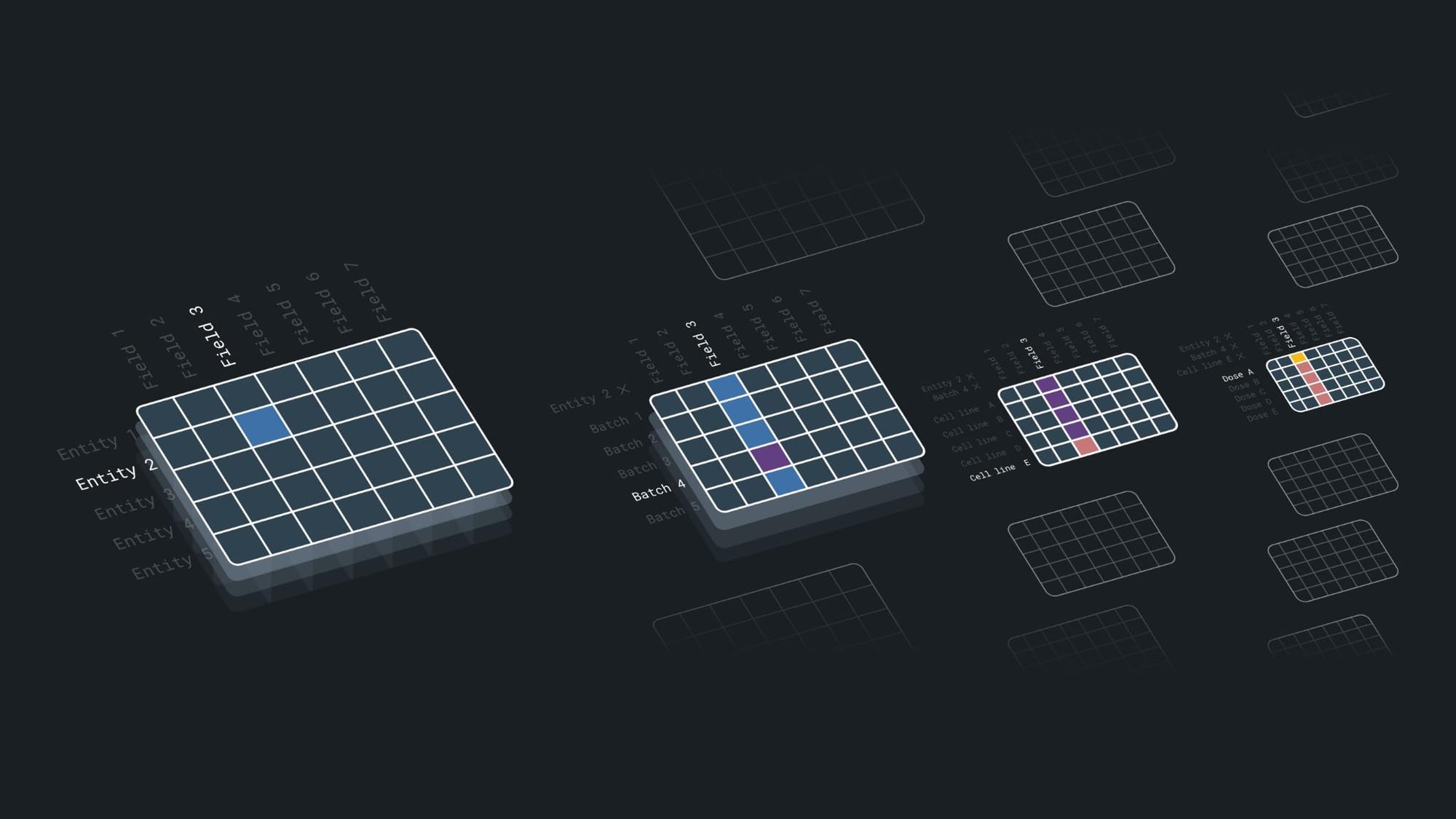
What does a better solution look like?
Companies need:
- Flexible data transformation tools that reformat metadata on the fly, adapting to instrument constraints
- Lineage-aware databases that automatically link past experimental contexts to new results
- Combinatorial-aware storage that handles multi-variable datasets without forcing manual restructuring
We've seen companies build some of these capabilities in-house, but few have been able to build comprehensive solutions that address all three needs.
Handling Different Data Formats
AI biotechs work with wildly different data types – from simple spreadsheet readouts to high-dimensional flow cytometry data to complex omics datasets. Each format has its own structure, visualization needs, and processing requirements. Most software forces scientists to become amateur data engineers, manually converting between formats before analysis can begin.
This creates several predictable problems:
Status tracking is rarely clear. Which datasets are complete? Which still need processing? Which contain quality issues? Scientists often resort to elaborate spreadsheet tracking systems or, worse, rely on memory to understand which datasets are ready for analysis.
Each data type also requires different visualization approaches to yield insights. While endpoint readouts like ELISA or absorbance assays fit neatly into spreadsheets, curve-based measurements - such as qPCR Ct values or dose-response assays - require tools that can capture the relationships between values over time or concentration.
Multidimensional datasets, like 40-color flow cytometry or spatial proteomics, introduce even more complexity, as each sample contains layers of interdependent measurements that don’t translate well into simple tables.
And at the most extreme end, omics data - like single-cell RNA sequencing or mass spectrometry - has no fixed dimensionality, making it difficult to store, structure, or analyze using conventional software. Most existing tools treat all experimental data as if it were neatly structured in rows and columns, forcing scientists to spend hours manually reformatting files before analysis can even begin.
An ideal software solution/stack should abstract this complexity by predicting, transforming and optimizing data formats automatically.
How should this work in practice?
- Format-aware software recognizes incoming data types and reformat them accordingly. A raw CSV from an NGS sequencer should immediately be structured into a database with lineage-tracked metadata - without requiring manual intervention
- Automated completion tracking that surfaces which datasets are ready for ML training and which need review
- Different experimental data types require vastly different visualizations. Flow cytometry data should open in a tool designed for multi-parameter gating and population analysis, not in a basic spreadsheet. qPCR Ct values should trigger standardized curve plotting tools without the need to manually adjust settings
- One-click data visualization triggers customized for different experiment types
These might sound like basic points but the context and requirements are complex. We've seen very few biotechs with software that checks all these boxes effectively. Most still rely on their scientists to manage format complexity manually.
Dataset Preparation & Analysis
AI models are highly dependent on structured, high-quality datasets. But preparing clean, structured datasets from messy experimental results remains one of the most labor-intensive aspects of AI-driven biotech.
The challenges here get very specific.
Linking experimental results back to their full history is often manual and error-prone. Result aggregation is inconsistent – scientists manually consolidate data across multiple platforms. Outlier detection happens slowly and manually, leading to dataset contamination. And dataset labels don't propagate correctly, breaking traceability between raw data and ML training inputs.
An effective software would include:
- Automated result correlation tools that link each readout to its full experimental history
- Intelligent result aggregation that preserves granularity while allowing high-level rollups
- Integrated outlier detection & review workflows that make it easy for scientists to approve/reject flagged anomalies
- Lineage-preserving dataset management that ensures labels correctly propagate into ML pipelines
With these capabilities, companies can rapidly convert experimental results into high-quality training data, dramatically accelerating their machine learning cycles.
Exception Handling & Automated Rerouting
In biotech, failure isn’t an option – it’s a given. Reagents degrade, instruments malfunction, samples vary in quality, and biological systems introduce inherent variability. A successful experiment might work 80% of the time. Most bio software ignores this reality. It assumes every experiment runs perfectly, leaving scientists to manually track and address failures.
This creates a cascade of inefficiencies.
Failed experiments get inconsistently documented, making it difficult to identify systematic issues.Without structured failure-handling workflows, companies rely on tribal knowledge – individual scientists deciding when to rerun experiments, often leading to inconsistent results, lost data, and wasted resources.
An ideal software would include:
- Event-triggered workflows that automatically rerun failed experiments based on pre-set conditions
- Rule-based rerouting (for example, "If 20% of plate wells fail, rerun with fresh samples; if 75% fail, trigger a reagent QC check")
- Exception libraries that grow over time, allowing teams to build reusable failure-handling rules
This approach transforms failures from disruptive events into routine parts of the research process.
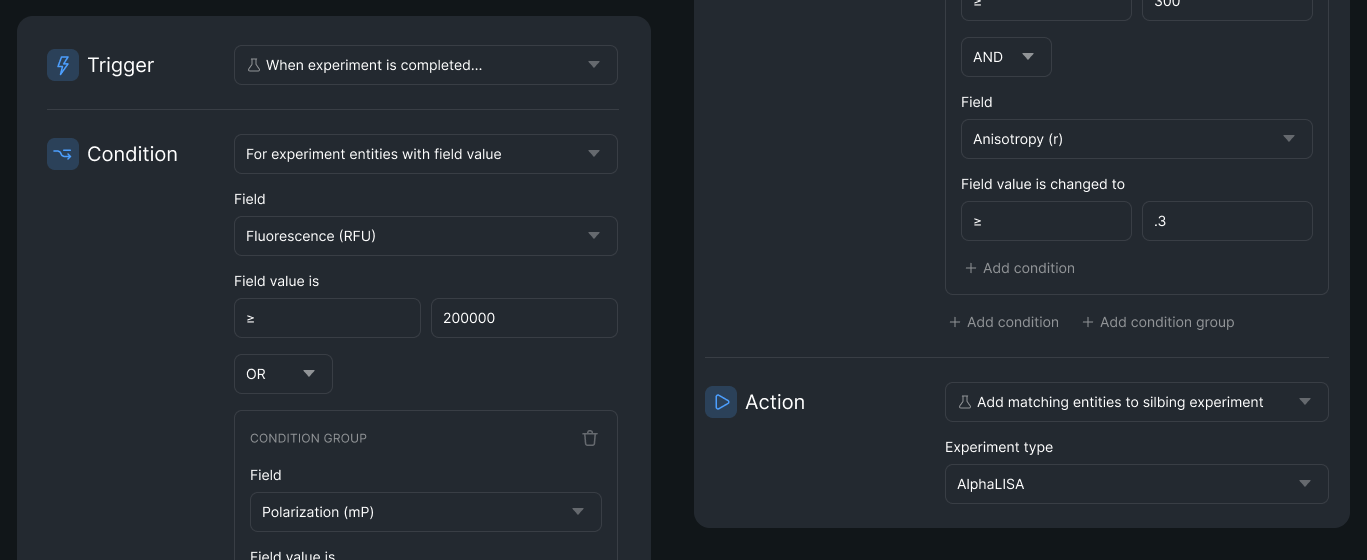
Coordinating & Monitoring Data Collection
Running a modern biotech involves coordinating dozens of instruments, hundreds of experiments, and thousands of samples – often with precise timing requirements. Yet most companies manage this complexity with a patchwork of calendar appointments, spreadsheet trackers, and tribal knowledge.
This approach breaks down as operations scale.
High-throughput experiments require precise coordination, but most lab software fails to integrate scheduling and monitoring in a way that reflects the true complexity of biotech workflows. A major challenge is that instrument operating systems rarely offer robust scheduling or real-time tracking capabilities, and the more advanced the technology - like spatial proteomics or single-cell sequencing - the worse the scheduling support tends to be. Many high-end instruments lack built-in monitoring tools, leaving scientists to rely on manual checks or external spreadsheets to track progress.
The solutions here involve better coordination at multiple levels:
- Project-wide coordination software that connects lab scheduling with experimental data tracking
- Run scheduling that integrates with instrument operating systems, allowing teams to manage experiments dynamically
- Automated monitoring dashboards that surface real-time data collection status
The right software ultimately turns data collection from a series of isolated events into a coordinated, observable process.
Why This Matters
After working with numerous AI-focused biotechs, a pattern has become clear: companies that treat data infrastructure as a strategic priority outpace those that view it as merely an IT concern. They recognize that data quality and accessibility directly impact the performance of their machine learning models.
The most successful organizations start by identifying their specific data bottlenecks. Is experimental data losing critical context? Are scientists spending excessive time converting between formats? Does dataset preparation create a bottleneck for machine learning? By targeting these specific pain points, companies can make focused investments that yield immediate benefits.
Equally important is recognizing that data infrastructure evolves alongside scientific capabilities. The systems that work for a small team running dozens of experiments per week will buckle under the load when that same team scales to thousands of experiments. Planning for this evolution from the start prevents painful transitions later.
Closing Thoughts
The competitive advantage for AI/ML biotechs doesn’t come from having slightly better algorithms – it comes from generating higher-quality data more efficiently and transforming that data into insights more effectively.
The companies that recognize this reality and invest accordingly gain a significant edge. They can iterate faster, test more hypotheses, and ultimately build more predictive models than competitors still struggling with fragmented data systems.
If you're building an AI-driven biotech, take a hard look at your data infrastructure. Are you generating and managing experimental data efficiently? Can you handle diverse data formats seamlessly? Do you have robust processes for dataset preparation? Have you built systems that handle exceptions gracefully and coordinate data collection effectively?
Your answers to these questions may determine your company's success more than any particular machine learning approach.
Kaleidoscope is a software platform for biotechs to robustly manage their R&D operations. With Kaleidoscope, teams can plan, monitor, and de-risk their programs with confidence, ensuring that they hit key milestones on time and on budget. By connecting projects, critical decisions, and underlying data in one spot, Kaleidoscope enables biotech start-ups to save months each year in their path to market.
