Software + science: what tools are R&D teams using?
A lot of our work at Kaleidoscope involves listening to people who work in life sciences: trying to understand what they’re doing versus trying to do, what’s working versus what’s breaking, and what problems they wish would go away. In this post, we summarize some of our learnings when it comes to the software that scientific teams are using to track their science. We hope that this post seeds some discourse around collective ways in which we can improve the state of the field. If anything here resonates with you, please reach out!
Summary points
- Flexibility vs rigidity: A common trade-off in scientific tracking software is between how flexible versus how prescriptive the tool is. While the former allows for nimble adjustments to account for the chaotic nature of early R&D, the latter ensures more robust data integrity. Enabling the right balance between these needs is a challenge in the field.
- Scalability and traceability: Commonly used software tools often fail in their ability to track work at scale. They become clunky, slow, and hard to use, and many of them are not equipped to map, manage, or query relationships between entities. This makes it very hard to preserve knowledge in a useful and searchable way, understand how an end result was generated, and maintain a holistic view of what’s going on in an organization at a given time.
- Multi-person or multi-team workflows: Capturing hand-offs across individuals or specialities is extremely difficult to do using today’s tools. There are no great systems for robustly tracking decision-making or effectively automating hand-offs between bench scientists and engineering teams. This problem is compounding due to the increasingly collaborative nature of R&D.
Well before we ever wrote a single line of code for Kaleidoscope, we spent a lot of time talking with numerous people working in bio. Bench scientists, computational biologists, software engineers, bioinformaticians, process engineers, chemists, data scientists, lab directors, team leads, CTOs — the list goes on. These individuals came from a range of types of companies, differing in things like size, stage, therapeutic modality, and business model.
The more we spoke with people, the more we learned about what tools they were using to track their scientific work, what they liked or disliked about them, and what was missing. Below, we summarize what we learned across >300 discussions, grouped by general type of tool. For more on why we started Kaleidoscope, check out our earlier post.
Electronic Lab Notebooks (ELNs) and Lab Information Management Systems (LIMS)
ELN and LIMS use is widespread; traditionally different tools, a number of providers offer unified solutions that borrow from both sets of capabilities. Some examples of commonly used software are Dotmatics, Benchling, Labguru, SciNote, and the PerkinElmer suite.
For ELNs, the main benefit is the value of having a flexible, purpose-built tool for bio, that provides powerful utility to the individual scientist, for things that scientists care about (e.g. being able to easily design a plasmid). Additionally, ELNs have enabled the move from paper notes to digital notes, which brings with it expected advantages such as better preservation and the ability to work with templates or shortcuts.
A commonly referenced drawback is that ELNs don’t solve for managing and tracking workflows across teams, since not all teams use ELNs; important hand-off or decision-capture is missed. This is an increasingly prevalent problem in a world where scientific experiments and results are not solely wet-lab-based, but might span numerous teams - like Data Science or Machine Learning - that work in separate software environments.
Additionally, ELNs are high cost and offer a poor product experience, especially at scale. These are not issues when a team is small and scientific throughput (e.g. number of experiments) is low. However, many ELN providers are extremely expensive at scale, while simultaneously becoming worse in terms of speed and ease of use of the product. Poor product experience leads scientists to move away from using their ELN as a day-to-day tool, and instead take hand-written notes and spend significant time every couple of months transcribing these to their ELN (for compliance purposes). This problem is especially pronounced for scientists that work with sensitive materials/samples, and therefore avoid bringing electronics to their lab bench.
In many ways, LIMS pros and cons are the reverse of those of ELNs. LIMS are highly structured systems for tracking data as opposed to notes: materials, reagents, and other entities. So while ELN use or structure often varies widely from scientist to scientist, the structure inherent to LIMS makes tracking information over time easier when operating at scale (e.g. for industrialized processes). Tying experimental throughput to things like inventory becomes possible when this sort of information is standardized and tracked in a LIMS.
The double-edged sword here is that science is inherently messy. While LIMS excel at tracking standard processes and work, such as closer to clinical stage, the reality is that a significant portion of R&D is less predictable and needs to adapt over time. LIMS adaptation requires specialized expertise, so reconfiguring a LIMS any time there is a change in something like the design of an experiment or the attributes being tracked, causes significant delay and pain for a bio company. Further, LIMS do not natively account for decision tracking, leaving a big gap between the 'how’ and the ‘why’ behind the science.
Google Docs or Sharepoint
A lighter version of ELNs, and one we hear about fairly often, is the use of something like Google Docs, as a way of keeping a flexible record of work over time. In addition to being light and easy to use, a main appeal of these docs is the synchronous editing capabilities, which some scientists love using for collaborating on work.
However, these can get unruly over time, either in size of a single doc, which becomes very slow to load especially when images are involved, or in number of individual docs, many of which are scattered across different Drives. Further, important context about the relationship of information, such as how two experiments were related or what several workflows may have led to a single result, is lost when spread across files. This problem is especially pronounced given the completely unstructured nature of these docs. There’s also the basic fact that these tools were not purpose built for scientists, so they lack the out-of-box capabilities that an ELN would have, such as design tools, the concept of bio-specific entities, or the ability to sign, witness, or lock experiments.
Spreadsheets (Excel, Google Sheets, etc.)
When the time comes to log information in a more structured way (experimental results, dates, batch numbers, references), scientists frequently work in spreadsheets. A significant advantage is the fact that set-up is minimal. Apart from naming columns and doing some basic formula work, scientists love being able to just put information into a tool like Excel and move on, and not worry about setting up and navigating a novel system. This especially makes sense in cases where a scientist is testing something out and wants as simple of a system as possible: quickly generate results, write them down, look at them, make note of adjustments, and generate new results.
Despite the draw of ease and familiarity, the cons of tracking work in tools like Excel are plenty. Some notable ones include:
- Scalability: Excel or Google Sheets are not meant to handle hundreds of thousands of rows or more, as is increasingly common in scientific work;
- Reliability of data: Excel is notorious for both creating errors, such as the classic gene-name-to-date autocorrect, as well as propagating them (e.g. when a formula is incorrectly configured or references are altered). Excel files can also be corrupted and are freely editable by others, which is dangerous when trying to rely on them as the source of truth;
- Configurability: coding macros for doing specialized analyses or manipulations is very inefficient and error-prone.
Additionally, the tradeoff with the ease of dumping data ad-hoc into an Excel file is the fact that this makes it harder to enforce consistency in how and what data is recorded, and makes collation/search across multiple sheets very difficult.
Powerpoint
Powerpoint is the worse version of spreadsheets. What started off as a tool to present results has permeated into many other areas, scientific tracking amongst them. Like with Excel, the Powerpoint format is very familiar to scientists and straightforward to use. In addition to the advantages of having a visual way of presenting things, since scientists present regular updates in Powerpoint, it also tends to be a consistent place where data ends up.
The problems arise when Powerpoint morphs from being the tool that’s used to highlight and present key figures, to the tool that’s used as a historical record of work. What starts off as a 20-slide .ppt file for lab meeting one week, quickly becomes a 100-page Frankenstein deck, with no clear link between how A led to B. Even worse than that is the tendency for scientists to create their own versions of a deck, quickly resulting in a slew of folders filled with confusing file names.
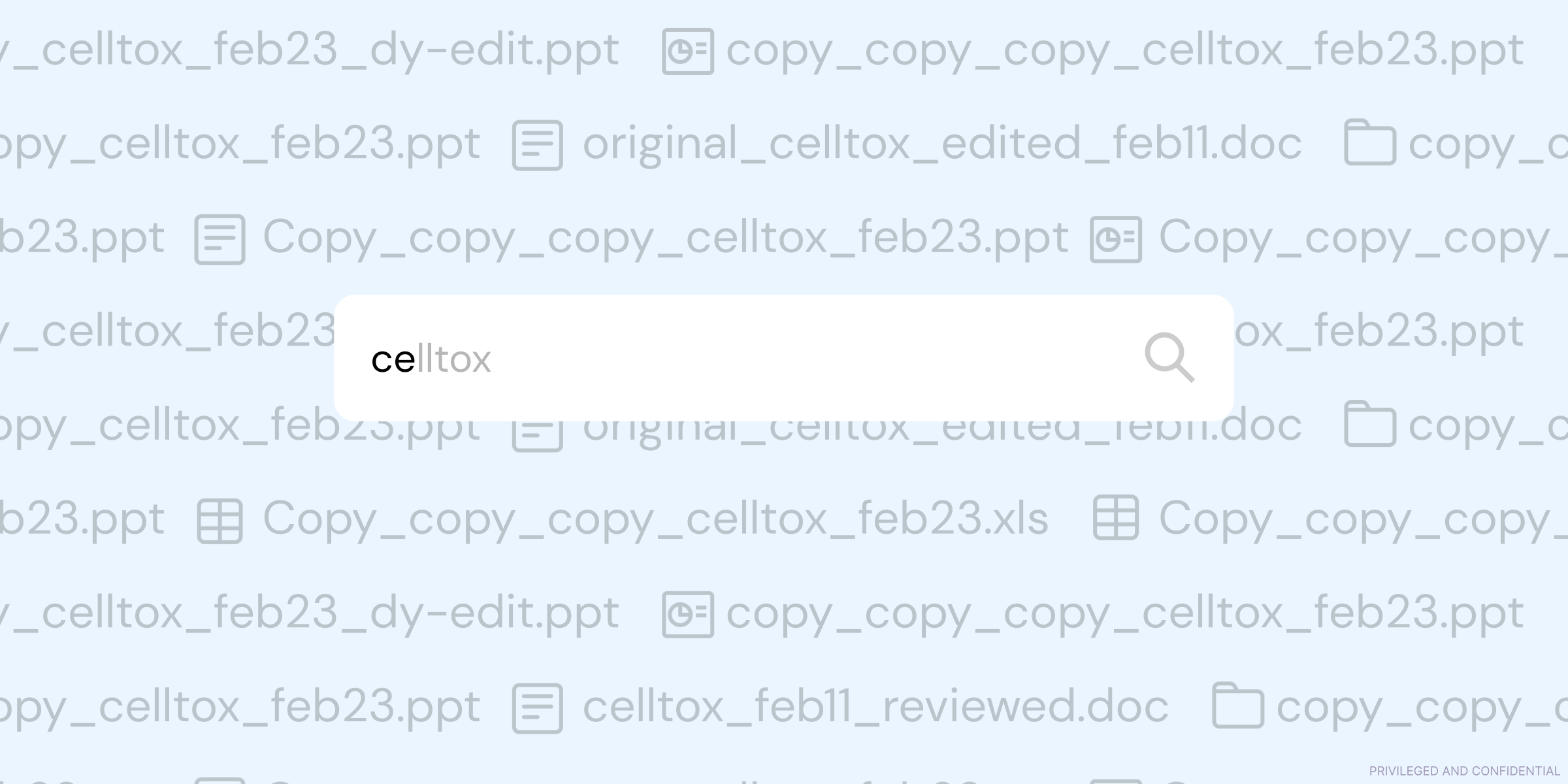
As you can imagine, we often hear about how this creates severe, compounding problems when it comes to auditability or traceability of results, as the link between data + output becomes murkier with each new iterative addition of slides.
Also, a large portion of scientific teams spin out and store .ppt files locally, meaning that when they move teams or leave the org, that knowledge is lost with them. Lastly, storing key results or decisions in Powerpoint makes it close to impossible to do any kind of programmatic analysis on that information, meaning large volumes of important information can’t easily be used to understand a system as a whole or potentially generate novel insights.
Traditional project management (PM) tools
We often hear about scientific teams trying to use more generic SaaS tools like Asana, Smartsheet, ClickUp, Monday, or Trello for organizing and tracking scientific work. The appeal for these stems from a few things:
- Product user interface: put simply, most of these products are much more pleasant to interact with than trying to hack together trackers in spreadsheets or older legacy software.
- The visual nature of the software: working with Kanban boards and Gantt charts can really come in handy when trying to zoom out and look at the bigger picture of things to do and timing of when to do them.
- And then there’s a larger category that we can think of as “modern SaaS actions”: things like the ability to assign tasks, set task dependencies, get notified of things, or integrate with other general software tools.
However, almost everyone we spoke with ended up unhappy with these traditional PM tools, to the point where they would sometimes need to resort to switching tools every quarter, in search of a better alternative. What we came to realize is that this discontent comes from the fact that the nature of R&D is fundamentally different than the use-cases for which traditional PM tools were built for. Tasks (the central unit in traditional PM systems) are ephemeral — they’re meant to be checked off and forgotten about. R&D, on the other hand, is the systematic building of interconnected knowledge over time; something that you do one day might become extremely important three months or three years down the line. And in a life sciences context, work centers around entities (compounds, samples, proteins, etc.) and processes (experiments, assays, or screens) rather than tasks.
Data linking was another common complaint — again, traditional PM tools were not built for tracking large amounts of data or for showing relationships between different data; in a scientific use-case, your tasks, projects, data, and results are all critical to your decision-making ability. Lastly, because the library of templates in an Asana, Monday, Smartsheet, or Trello was not built with science in mind, it takes a massive effort to set up a complex, custom scientific workflow that can be managed, tracked, and automated effectively. And since the body of ‘projects’ grows rapidly in a scientific context (you might be running many thousands or more experiments a week), attempting to hack something together in a traditional PM tool almost always breaks at scale.
Jira
In cases where we spoke to teams with an engineering background, Jira was often the tool of choice in place of an Asana or Trello. The ticketing system works well when it comes to tracking work as an Engineering team, and is a familiar one to people from a software background (for this post, we’ll ignore the grumblings that engineers broadly have about Jira). This was especially consistent with teams where Engineering is a separate, independent unit that rarely interacts with the scientific teams.
However, this is rarely the case in bio, where the value increasingly lies at the intersection of engineering and science. While Jira is great for agile software development, it does not have connections to the chemical, biological, etc. data being managed, nor does Jira come with functionality for evaluating that data or making decisions on it (e.g. compound or lineage awareness). The end result is that teams trying to coordinate across engineering and bench science roles still end up having to hack together systems that bring the two together, when trying to track scientific progress or make key organizational decisions (often, this ends up being in-house tooling — more on that below).
Notion
A tool that’s come up in a number of instances (especially for smaller or newer teams) is Notion, its main selling points being cost, product, and flexibility. Notion is very affordable for teams to get up and running with, which goes a long way for lean teams just getting started. The clean UI and modern design principles offer stark contrast and welcome respite from the software that most scientists are used to trying to wrangle with. Notion is also built with flexibility in mind, meaning teams can have free reign with how they structure their pages/knowledge base, using the tool’s various building blocks and templates.
However, this flexibility tends to be the main drawback for teams that have grown to more than several people. Complete flexibility can feel overwhelming at times and makes it hard to enforce best practices — eventually, people start diverging in how they structure or log things, which presents major challenges to a biotech trying to maintain a cohesive internal system. Similarly, Notion’s flexibility means that data integrity can quickly become compromised, as rules around data types, input standards, or metadata tracking are very hard to enforce from a product perspective (we previously wrote about the importance of tracking metadata).
This problem is compounded by the fact that although Notion has many out-of-box templates, they’re not built with R&D in mind. It takes significant effort to try and build these in-house, and even if built, simply attempting to maintain a robust system (let alone trying to extend it to bio-specific or internal tool integrations/automations) is too failure-prone to be worth it. When you combine this with the fact that Notion slows down significantly at a meaningful scale, you’re left with a system that most people are forced to move away from relatively early on as a company, at least for tracking scientific data/work.
Custom in-house build
For companies with experience dealing with the many hardships above, a common end result is that they decide to build their own internal software for tracking work. Naturally, the main advantage here is the bespoke nature of the approach; rather than hacking together numerous other tools built for very different use-cases and hoping that they’ll work for a science flow, you get to build a tool that does what you need it to do, for your specific context. A large number of companies we spoke to (big and small) went down this route or are considering it, because of the pressing need for better software to do science.
The decision to do this is not an easy one. Building good software that works how you need it to, especially when it comes to managing complex work and data flows, is hard. It takes significant engineering resources — often multiple engineers’ time over multiple years. Maintaining these tools then presents an entirely different challenge: how do you ensure that the systems don’t break at scale or over time, especially as the information contained within grows more valuable? On top of that, going from “we have a tool that works” to “we have a tool that works and that can be used by the (often non-technical) people that need it” is non-trivial.
Effectively, these Life Sciences companies end up trying to run small software companies from within, which in addition to being extremely difficult, is both far from their core competencies and a massive distraction/resource sink from the work they should be doing. The end result is often years of work and millions of dollars spent on poorly designed, hard-to-use software. When we talk to experienced teams or leaders who’ve spearheaded this work before, by far the most common answer we hear is “I wish there was off-the-shelf software for this” or “I never want to do this again”.
Conclusion
While the pace of scientific advancement over the decades has been staggering, the pace of software development to serve scientists has been lacking. We believe that there is a massive opportunity to bolster the work done by scientific teams everywhere, and that the answer lies in applying a deeply empathetic product engineering + design lens to the context of the work being done. Work that is foundational to our understanding of the world around us: to our understanding of health and disease, ecological preservation, and technological advancement more broadly.
If you want to chat more about anything we wrote, or you’re interested in finding a way to work together, let us know!
